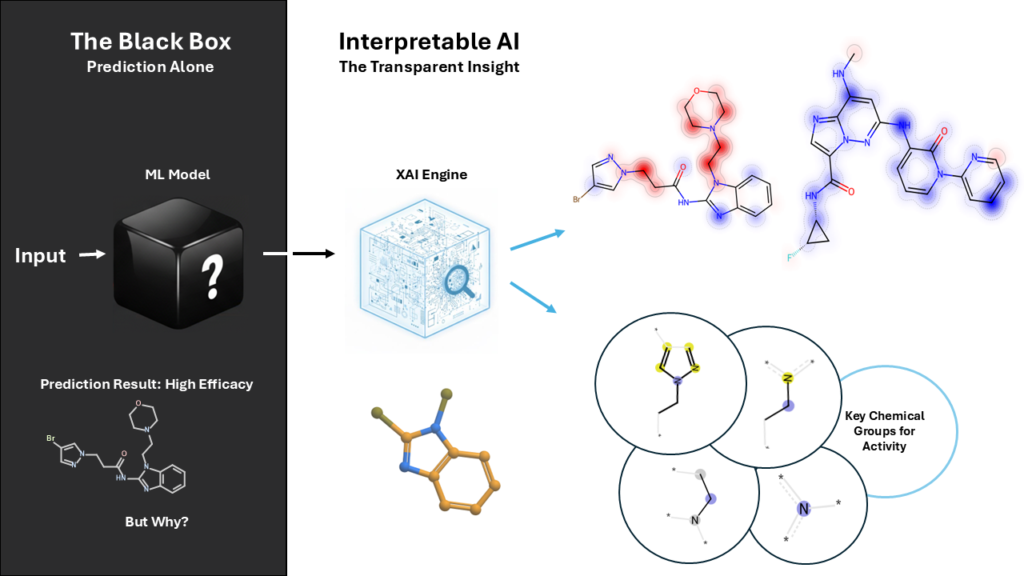
Technologies for drug discovery
- Ligand based
- Structure Based
- AI / ML
Ligand based
Multi parameter lead optimization (MPO)
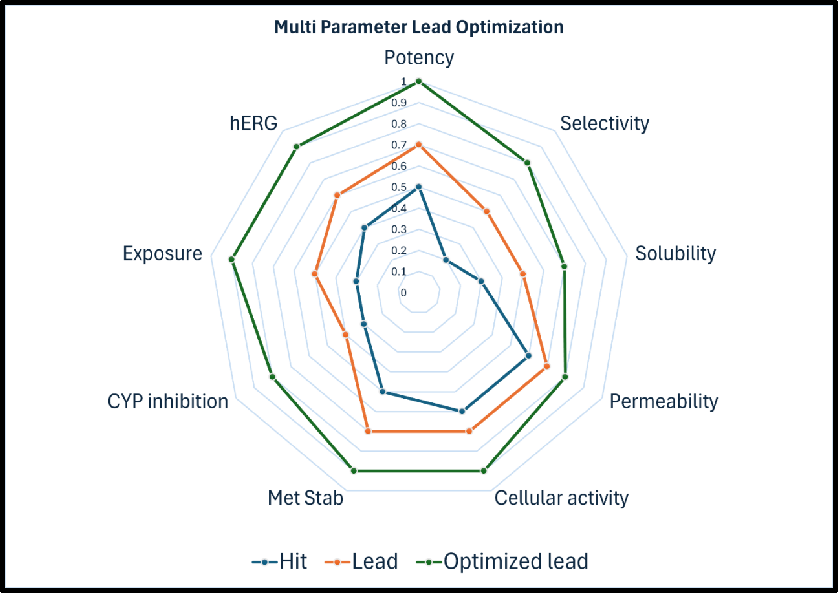
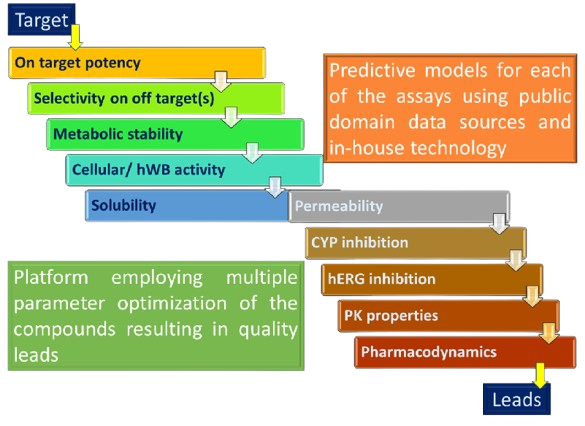
- ML/DL-based models predict DMPK parameters and help prioritize syntheses
- Multi-parameter optimization driven effort to realize the evolving TPP in the series
Virtual Screening
Large collection of commercially available compounds for screening
- Structure based virtual screening (SBVS)
- Ligand based virtual screening (LBVS)

Large collection of commercially available compounds for screening
- Structure based virtual screening (SBVS)
- Ligand based virtual screening (LBVS)
Ultra Large Library Generation
Generative Chemistry
- Structure-aware molecule generation
- De novo design
- Superstructure generation
- Scaffold decoration
- Motif extension
- Linker generation
- Scaffold morphing
- Pocket-conditioned diffusion models
- Property-guided molecule optimization
- LLM-driven chemical ideation
- Evaluation and filtering
Density Functional theory (DFT)
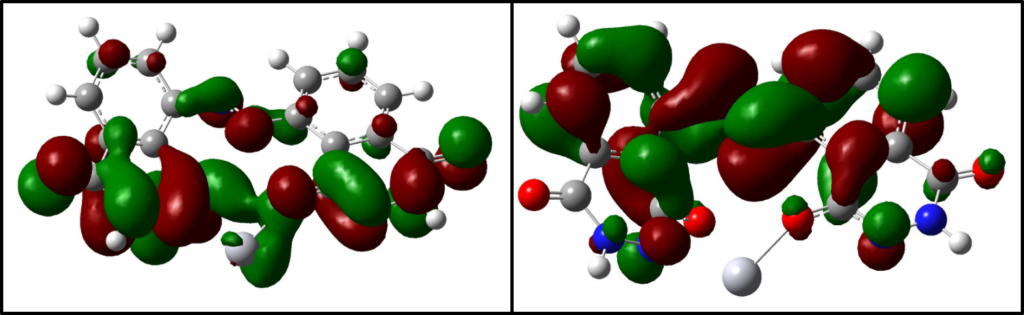
- Lowest energy conformation of ligands
- Understand electronic properties and reactivity of a compound through Frontier Molecular Orbitals (FMO) approach
- Novel isostere identification through electronic properties
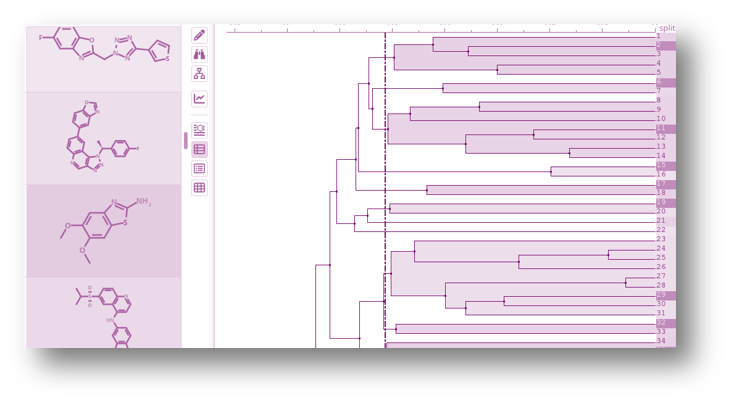
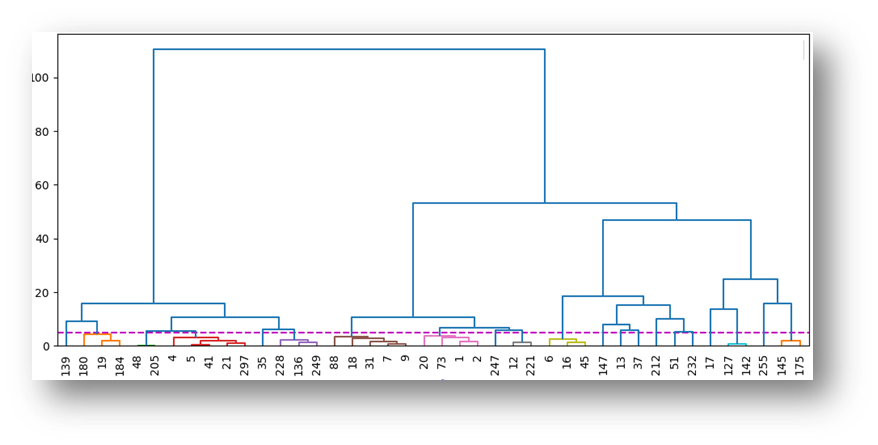
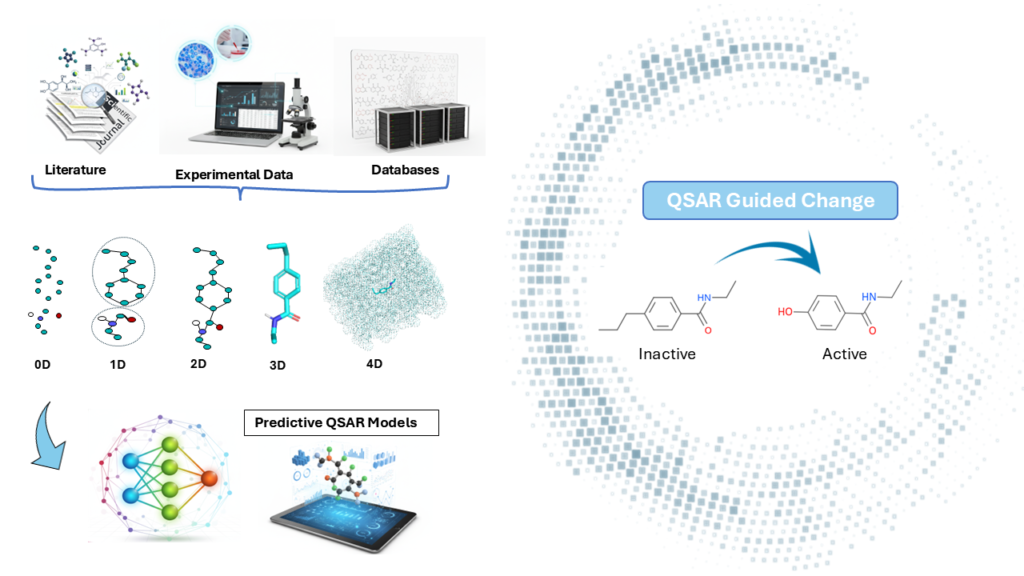
Structure Activity Relationship and Molecular Matched Pairs
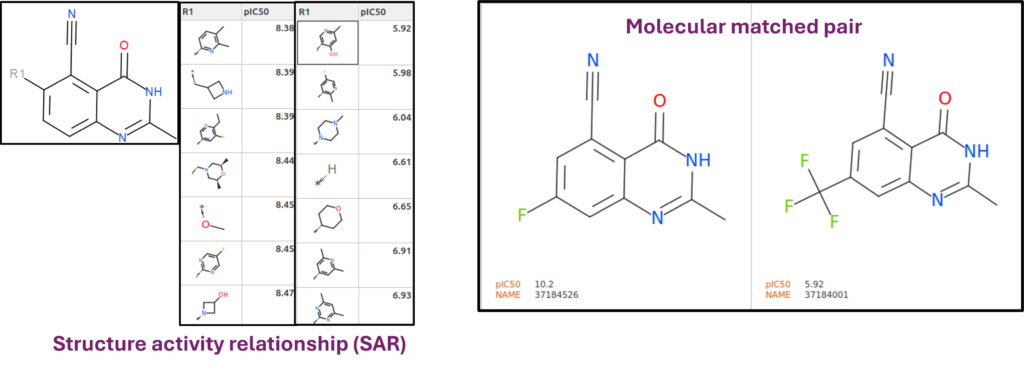
Structure activity relationship (SAR) showcased through a variation of R group
Structure Activity Relationship and Molecular Matched Pairs

Structure activity relationship (SAR) showcased through a variation of R group
Conformational search
R-group Analyses

Structure activity relationship (SAR) showcased through a variation of R group
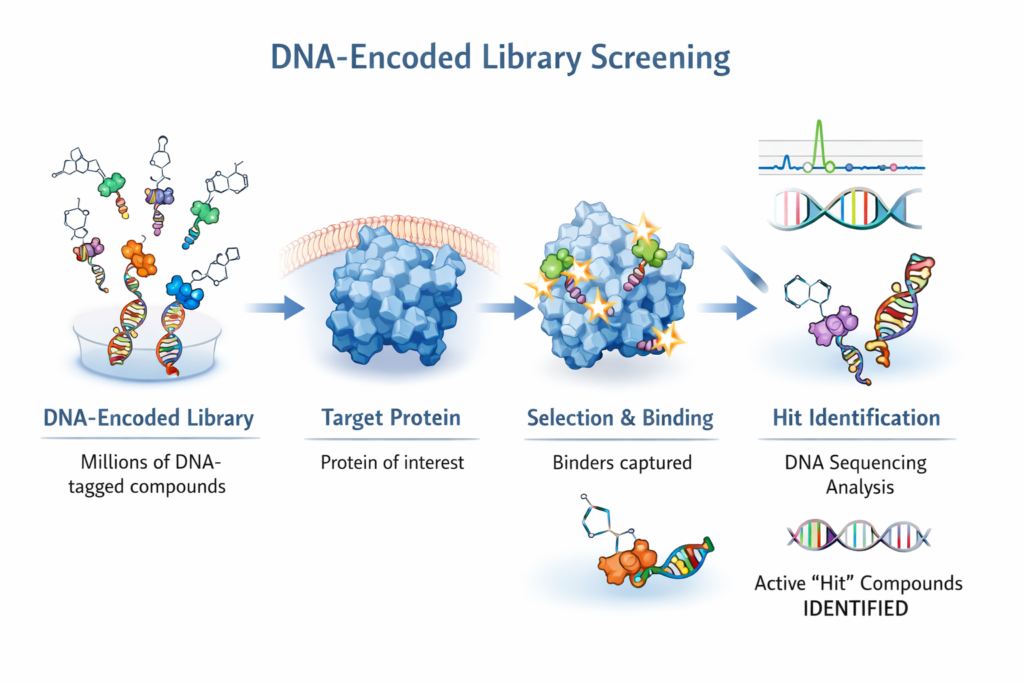
DEL/ASMS Screen Planning
- DEL/ASMS screens expand druggable target space by offering chemical starting points
- No, we don’t do DEL/ASMS screens but we can plan it for you and connect you appropriately
- This is the workflow we adopt:
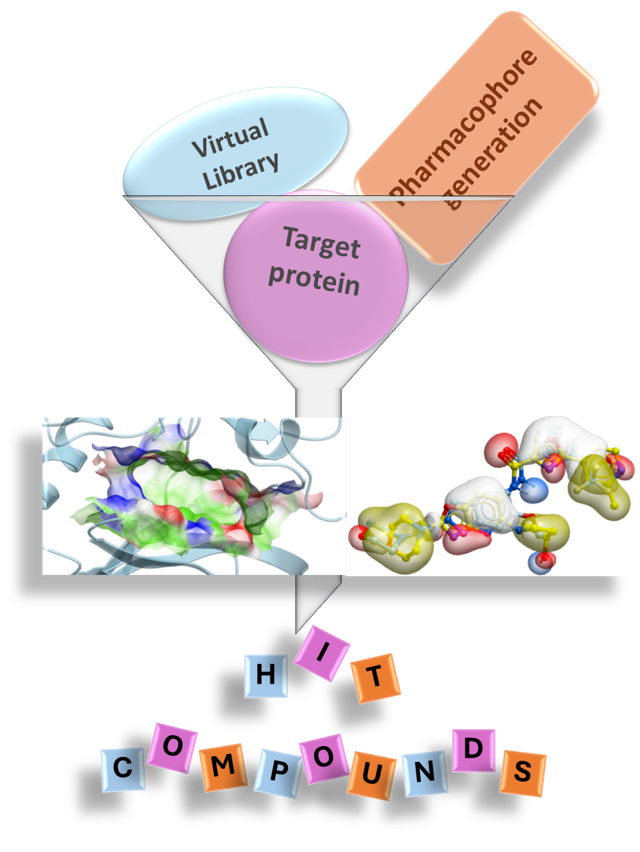
Pharmacophore

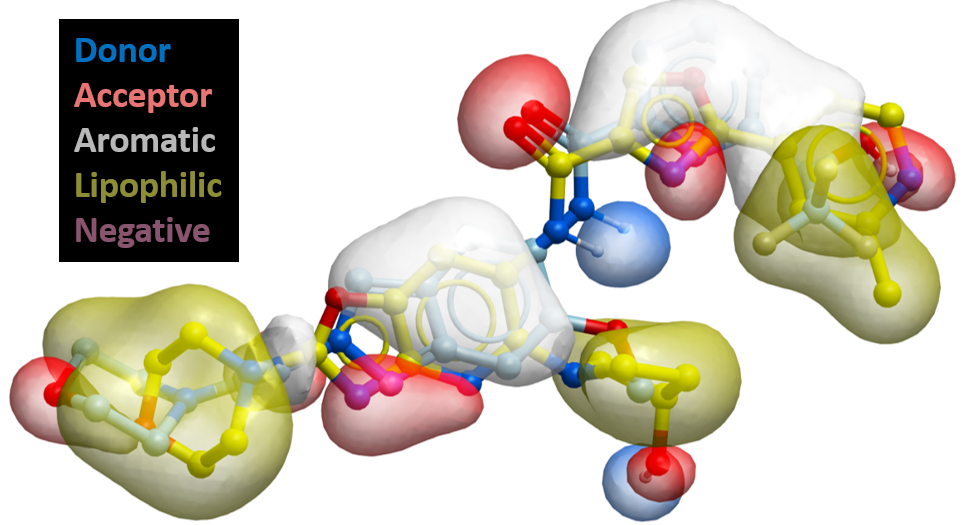
- Consensus and structure based pharmacophores
- 3D QSAR
- Screening against large library of compounds for a target protein (Ligand based virtual screening – 300M+)
Structure based
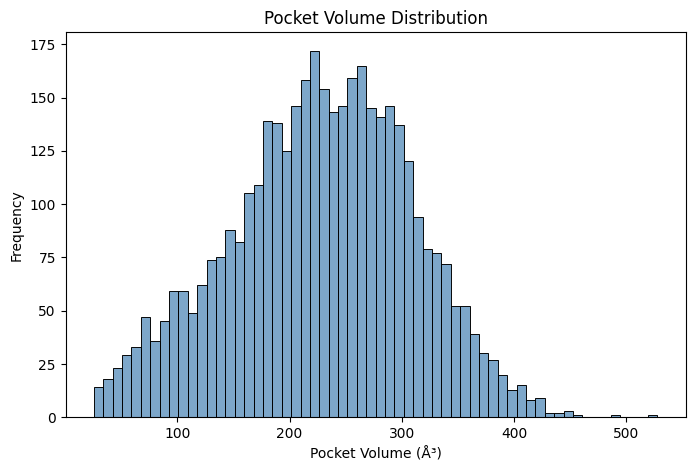
Pocket Druggability
- Pocket detection and segmentation
- Druggability scoring and ranking
- Binding site classification
- Ligandability prediction
- Dynamic pocket analysis
- Cross-target pocket comparison
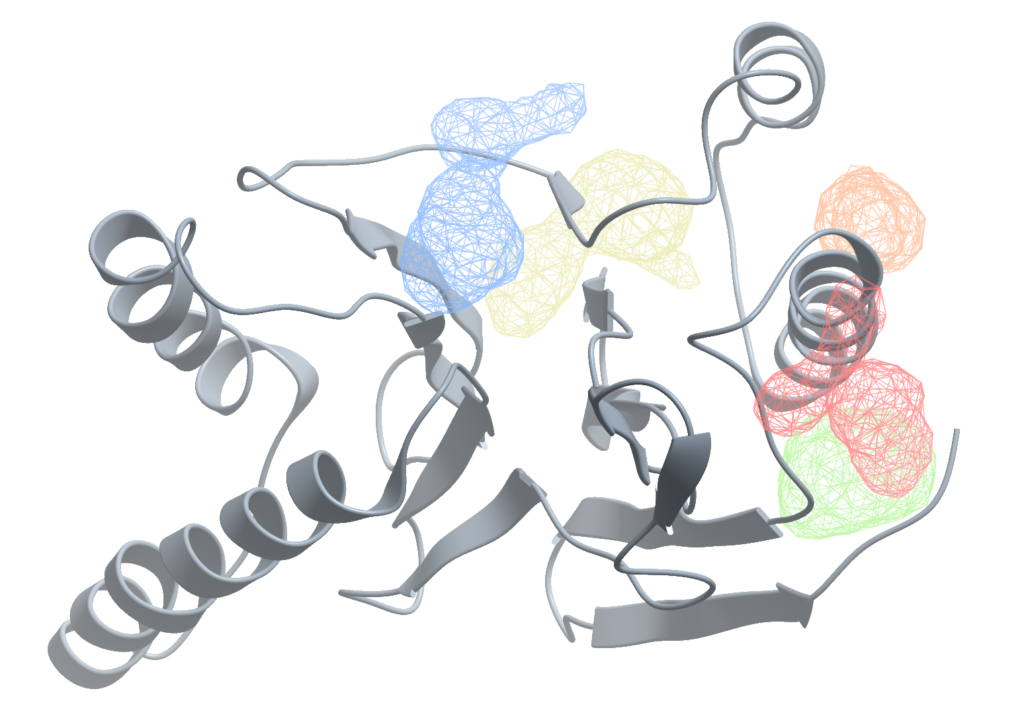
InteractogramTM
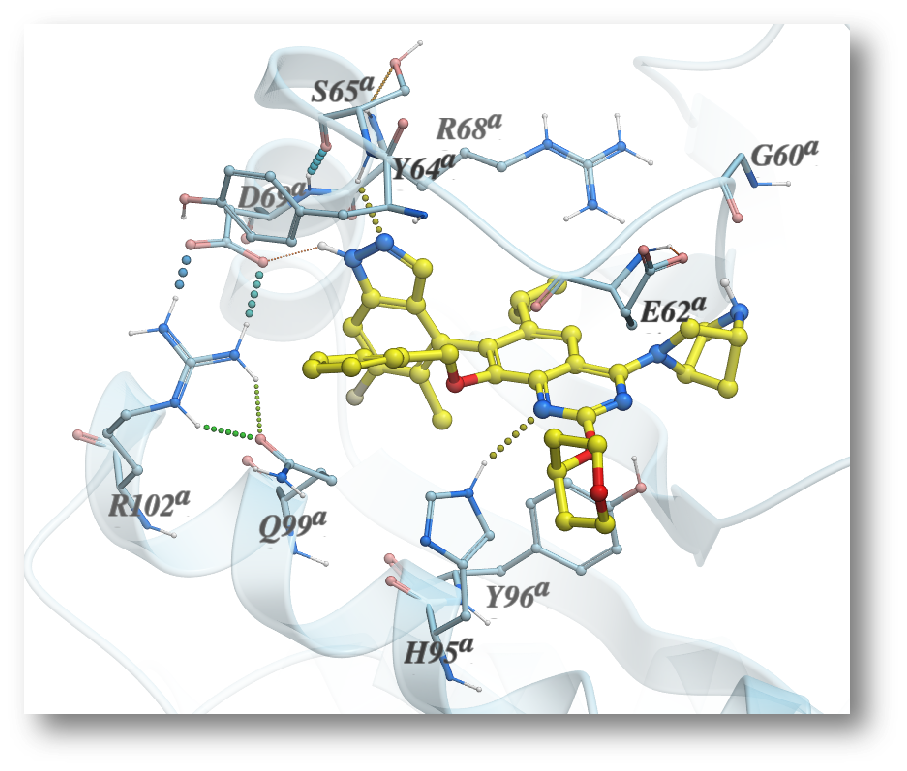
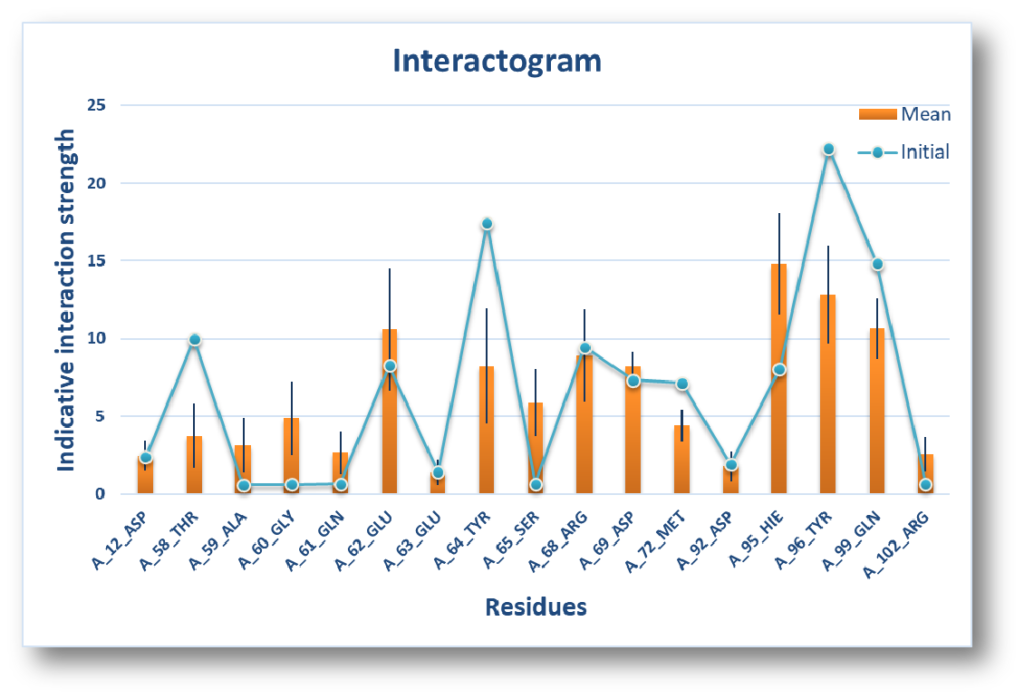
InteractogramTM : Where the ligand meets the interaction hotspots – IFP
HydrogramTM
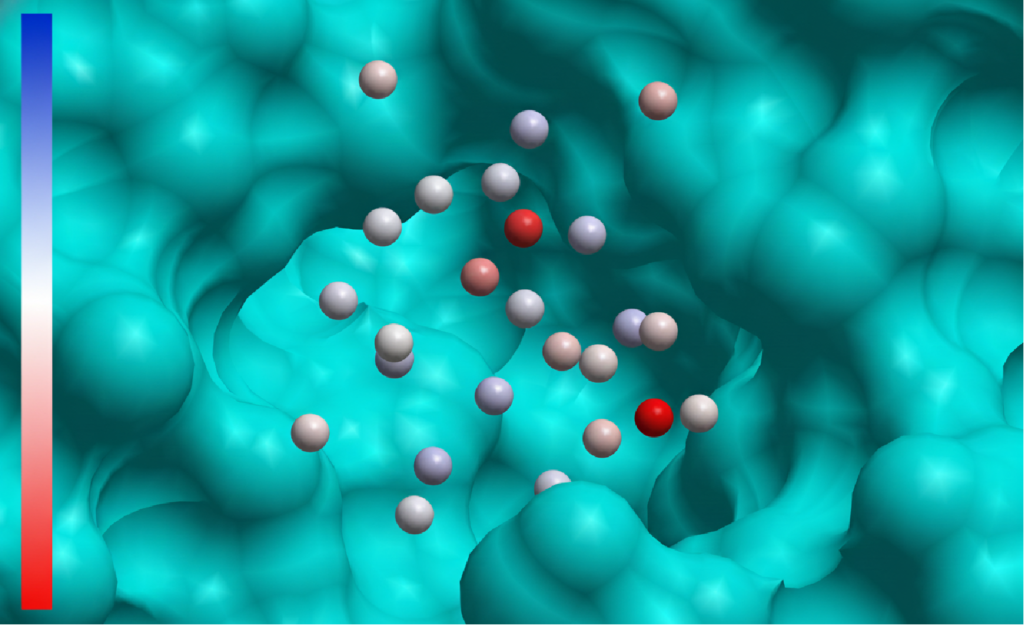
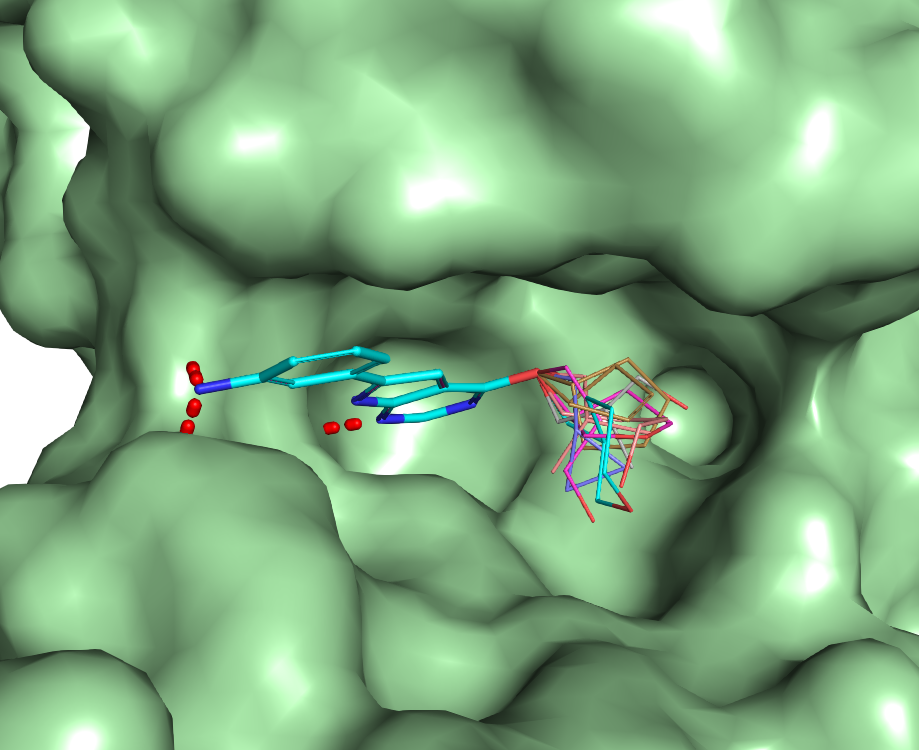
Identifying structural water sites in the binding pocket
Molecular docking
- Ensemble docking grid to account for the flexibility of the binding site residues
- Covalent, peptide and protein-protein, RNA docking
Covalent Docking
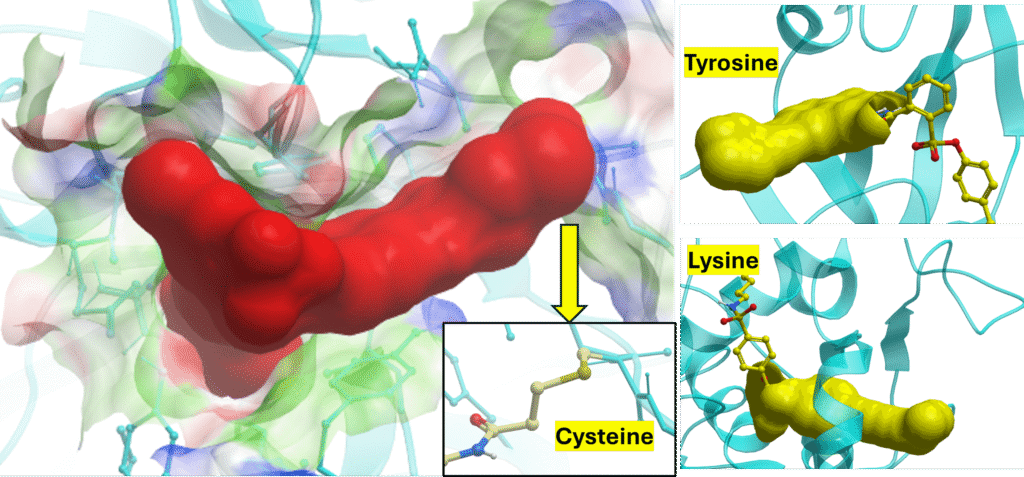
- Targeting suitable residues with covalent warheads
- Trap a range of reactive residues – Cys, Tyr, Lys, Ser, His
- Warhead electrophilicity assessment and modulation
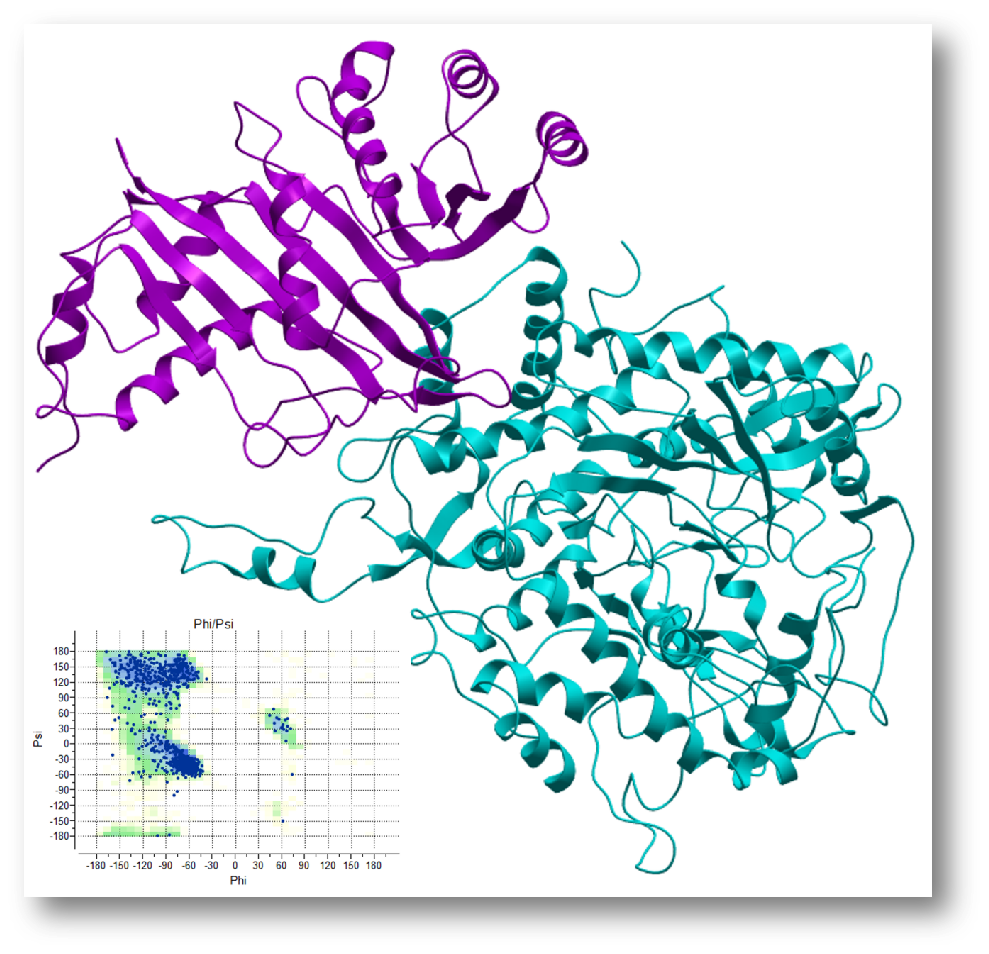
Protein-Protein Docking
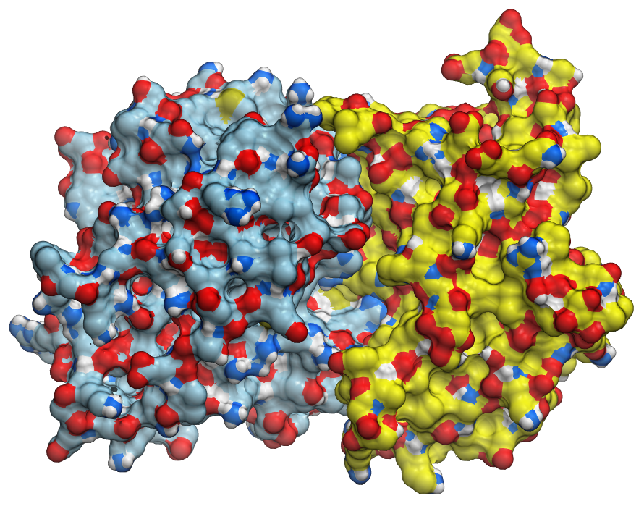
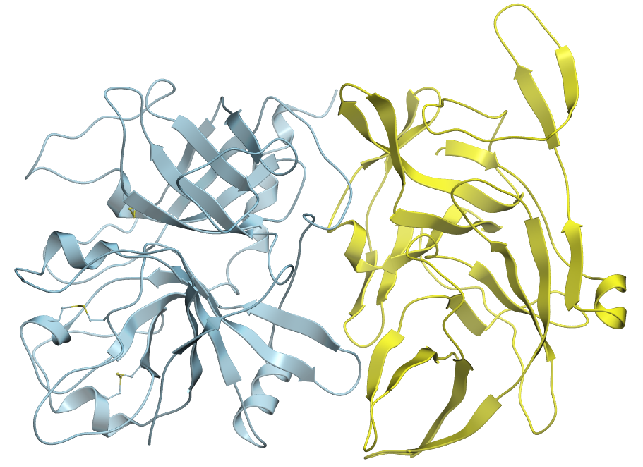
Identify probable binary complexes using energetics, clustering and ML
Virtual Screening

Large collection of commercially available compounds for screening
- Structure based virtual screening (SBVS)
- Ligand based virtual screening (LBVS)

Large collection of commercially available compounds for screening
- Structure based virtual screening (SBVS)
- Ligand based virtual screening (LBVS)
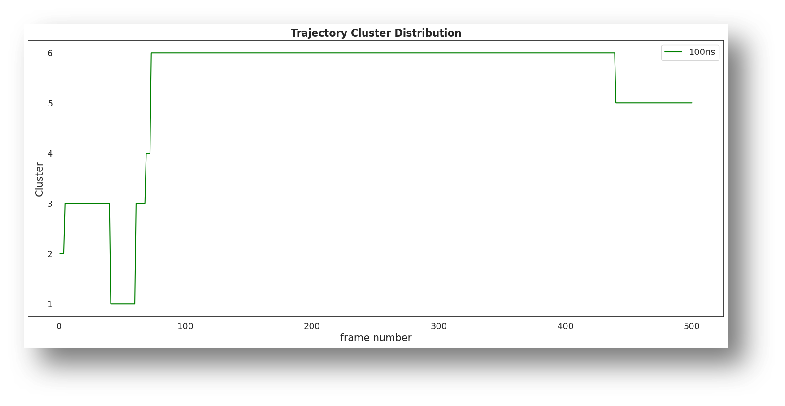
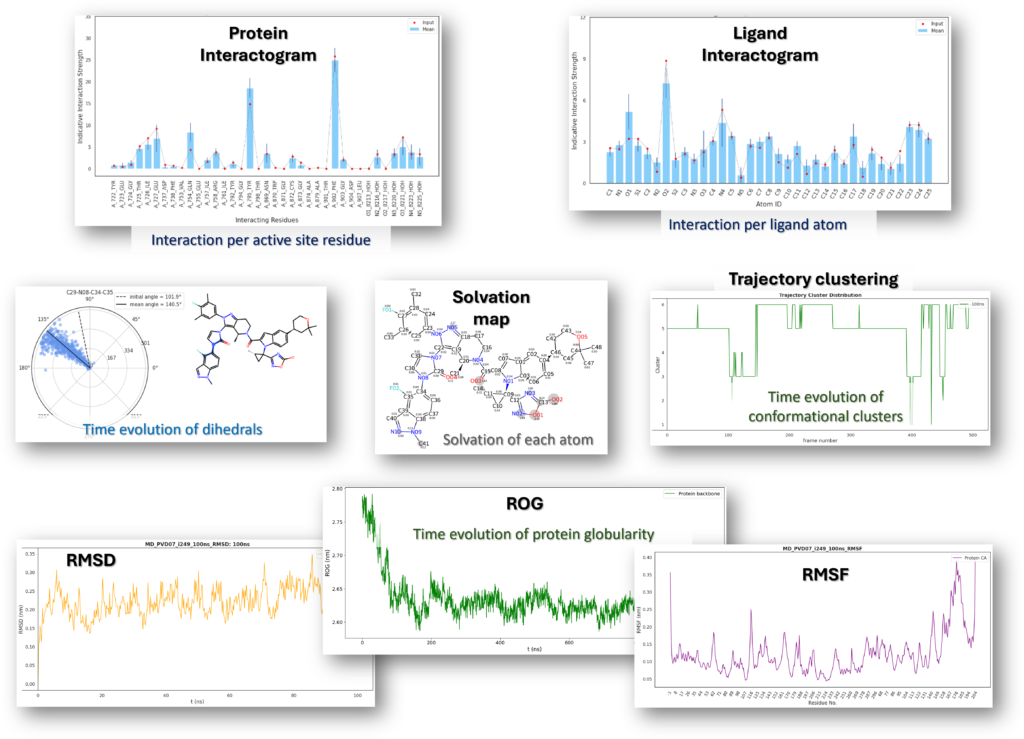
Molecular Dynamics Simulations
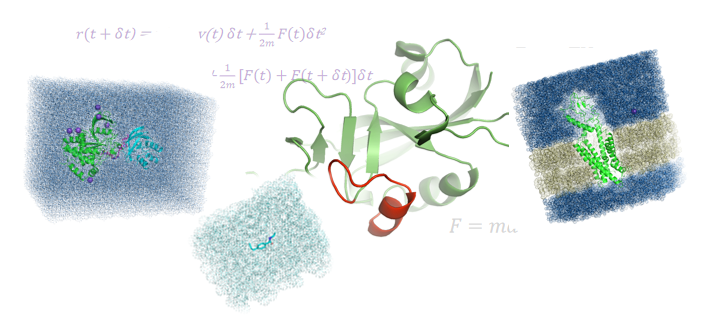
Physics based simulations to identify preferred protein states to yield novel designs
RNA Docking
- RNA structure preparation
- Binding site identification
- Ligand docking
- Protein–RNA docking
- Scoring and ranking
- Conformational flexibility
- RNA-quadruplex modeling
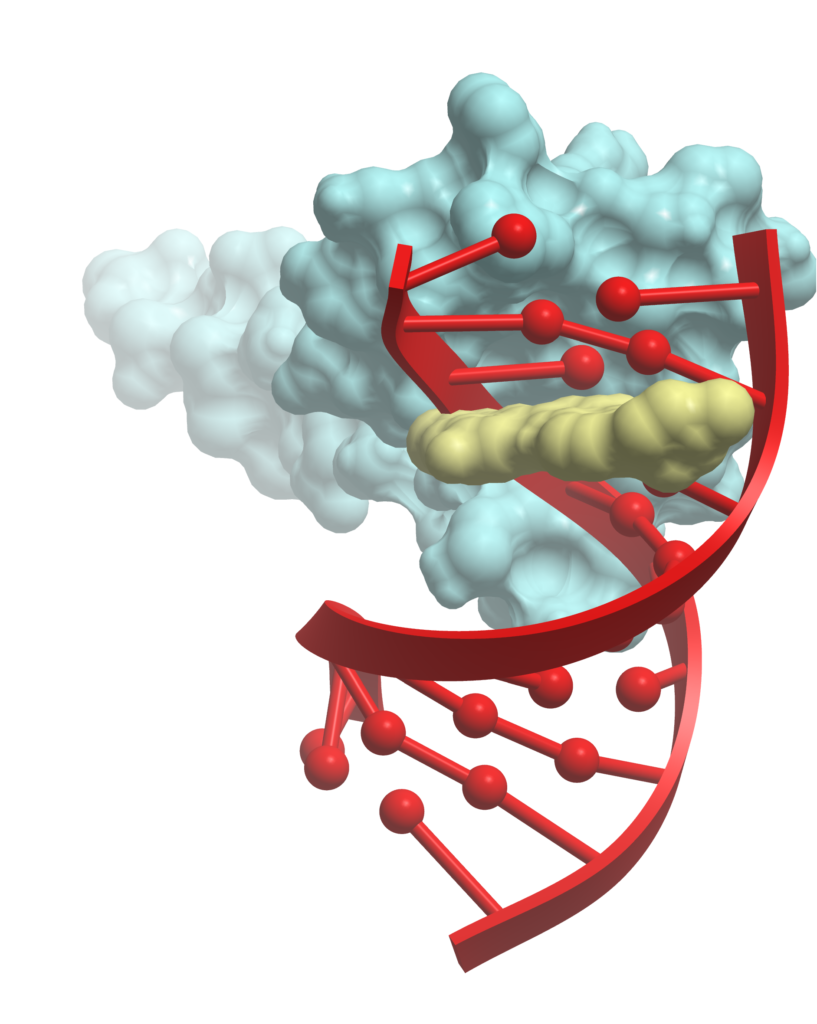
PROTAC Modeling
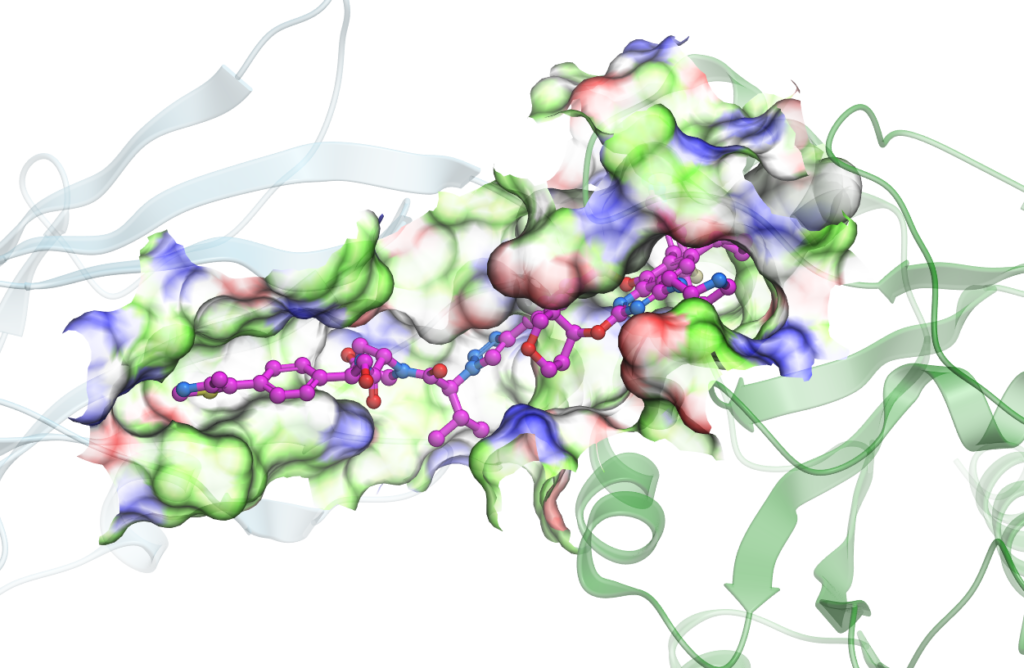
- Ternary complex generation for novel PROTACs
- Design and prospective modeling of PROTACs
Receptor Flexible Docking

- Ensemble docking grid to account for the flexibility of the binding site residues
- Covalent, peptide and protein-protein, RNA docking
Work in progress…
Peptide design

- Proteins with flat and shallow binding pockets
- Targeted through linear/cyclic peptide design
- Improving the druggable target space
- Deep Learning based peptide design
- Macrocyclic peptides
- Interaction hotspots using XAI
Fragment based drug discovery (FBDD)
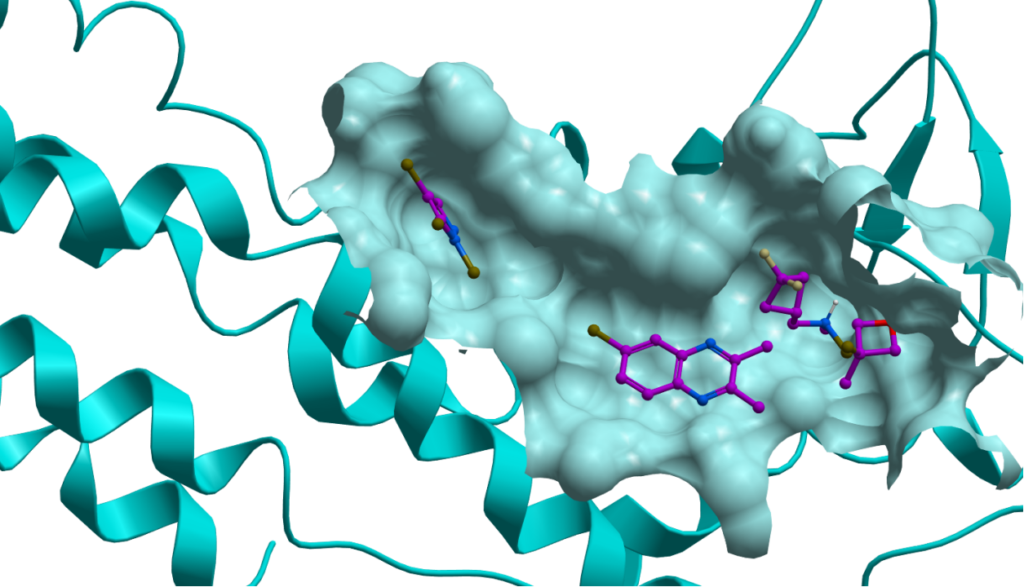
- Identifying interaction hotspots in the binding site
- New ligand designs through hybridization of fragments
- In-situ ligand editing
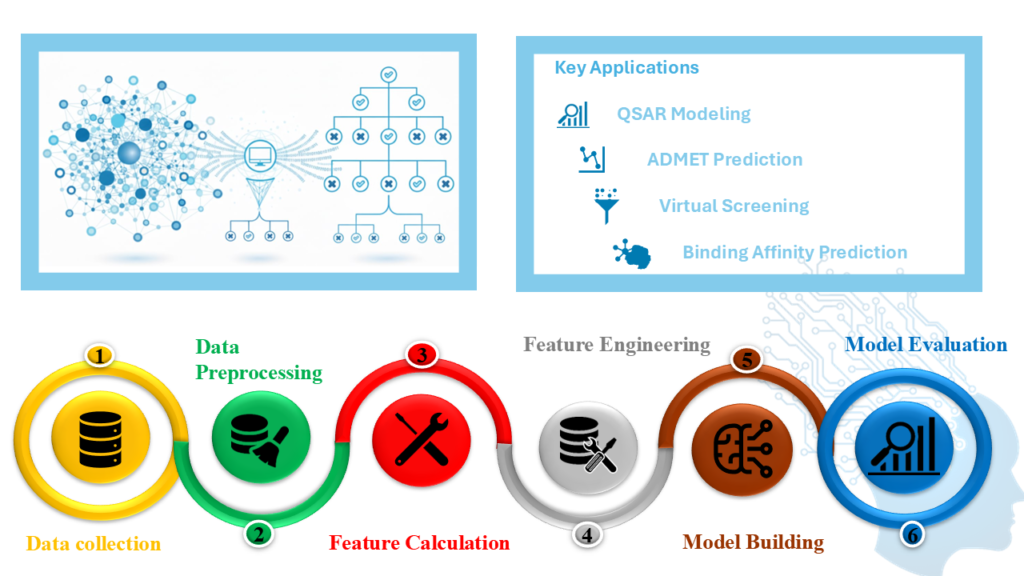
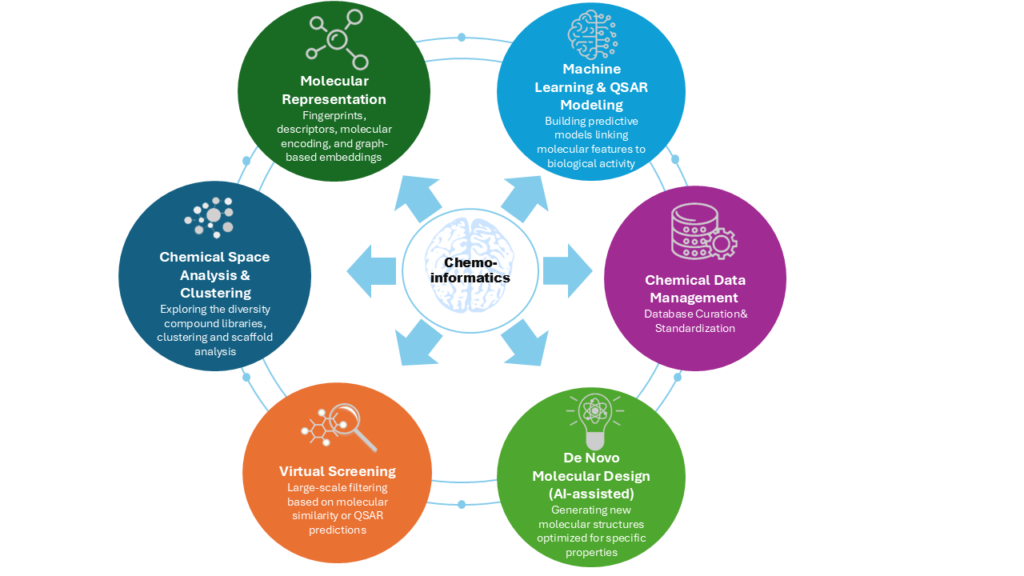
AI/ML
Deep Learning
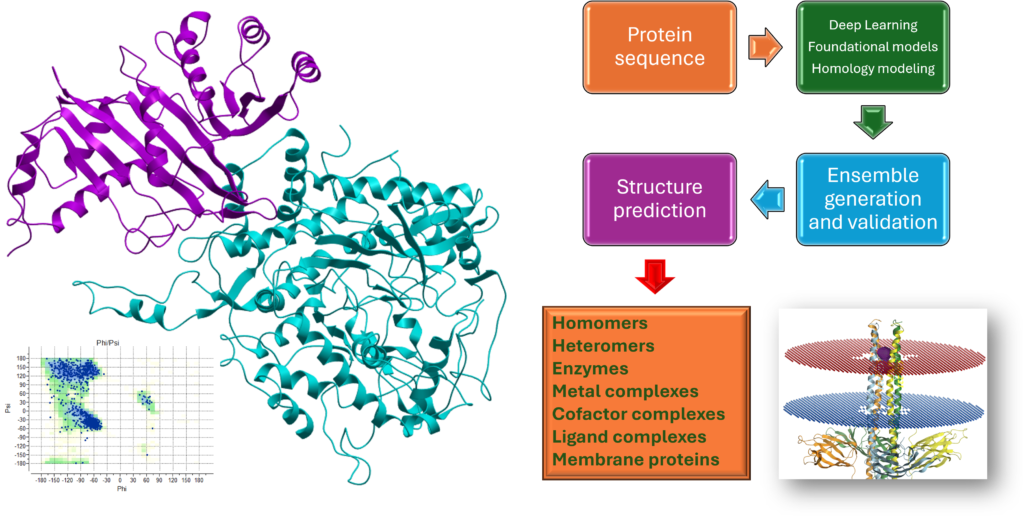
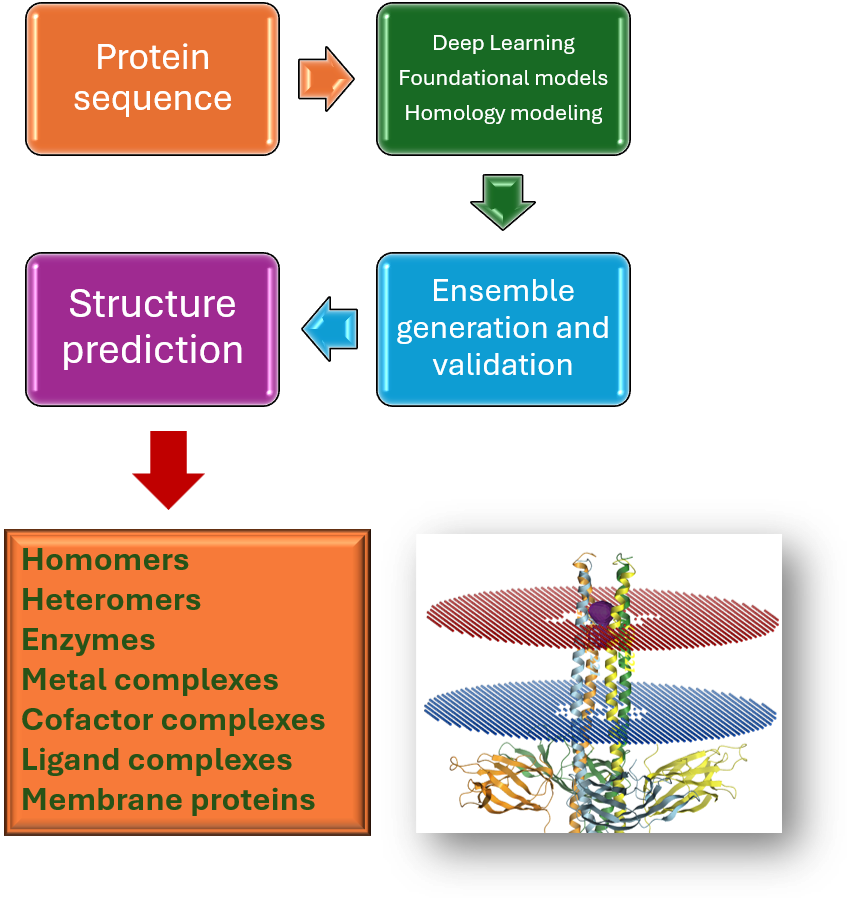
Structure prediction
- Sequence → structure prediction
- Multimer / homomer / heteromer generation
- Peptide generation
- Complexes with ligands / nucleic acids
- Conformational Ensemble Prediction
- Mutagenesis experiment
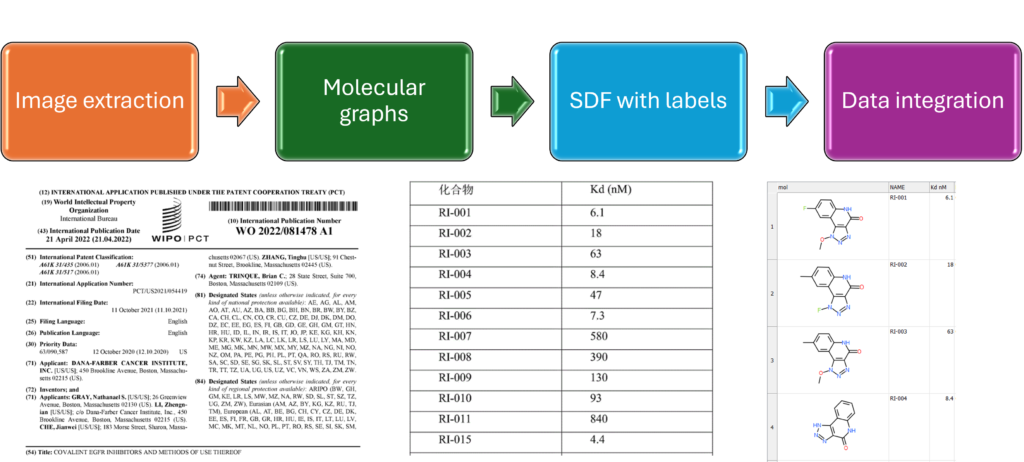
Patent digitization
- Structure recognition and extraction
- Data integration and cross referencing
- Text and data extraction
- Dataset from patents
Molecular designs
- Structure-aware scaffold decoration
- Multi-anchor decoration
- R-group generation with variable size
- Pocket-conditioned generation
- End-to-end diffusion-based generation
Property prediction
- ADMET and drug-likeness prediction
- Tautomer / protonation state enumeration
- Micro-pKₐ Prediction
- Binding affinity estimation
- Building DL models
Generative Chemistry
- Structure-aware molecule generation
- De novo design
- Superstructure generation
- Scaffold decoration
- Motif extension
- Linker generation
- Scaffold morphing
- Pocket-conditioned diffusion models
- Property-guided molecule optimization
- LLM-driven chemical ideation
- Evaluation and filtering

Knowledge Graphs
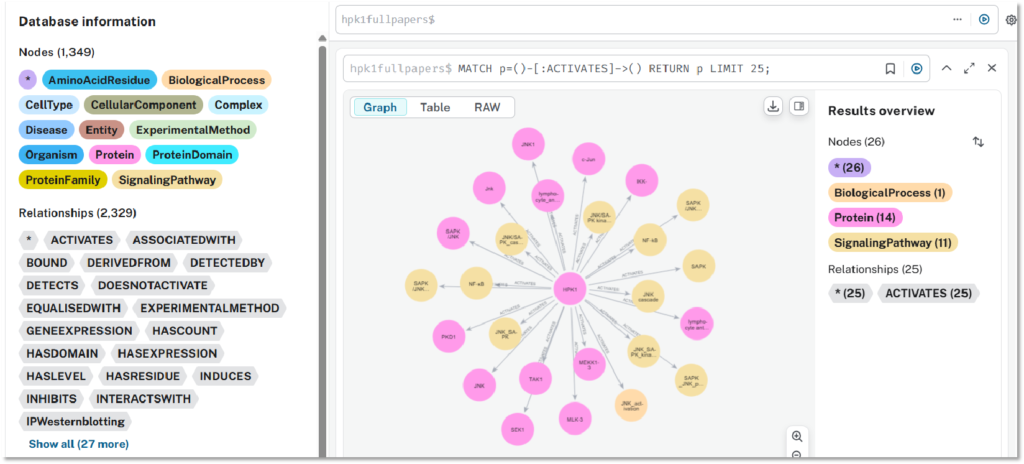
Refining vast information into decisive knowledge graphs